How to build a Metropolis-Within-Gibbs sampler?#
Gibbs sampling is an MCMC technique where sampling from a joint probability distribution \(\newcommand{\xx}{\boldsymbol{x}}\newcommand{\yy}{\boldsymbol{y}}p(\xx, \yy)\) is achieved by alternately sampling from \(\xx \sim p(\xx \mid \yy)\) and \(\yy \sim p(\yy \mid \xx)\). Ideally these conditional distributions can be sampled from analytically. In general however they must each be updated using any MCMC kernel appropriate to the conditional distribution at hand. This technique is referred to as Metropolis-within-Gibbs (MWG) sampling. The idea can be applied to an arbitrary number of blocks of variables \(p(\xx_1, \ldots, \xx_n)\). For simplicity in this notebook we focus on a two-block example.
import jax
import jax.numpy as jnp
import jax.scipy as jsp
import blackjax
from datetime import date
rng_key = jax.random.key(int(date.today().strftime("%Y%m%d")))
The Model#
Suppose that \((\xx, \yy)\) are drawn from a multivariate normal distribution
$$ (\xx, \yy) \sim \operatorname{MvNormal}(\boldsymbol{0}, \boldsymbol{\Sigma}). $$
The corresponding log-probability function is implemented below.
def logdensity_fn(x, y, Sigma):
"""
Log-pdf of ``(x, y) ~ MvNormal(0, Sigma)``.
"""
z = jnp.concatenate([x, y])
return jsp.stats.multivariate_normal.logpdf(
x=z,
mean=jnp.zeros_like(z),
cov=Sigma
)
# Specific example with x.shape == y.shape == (2,)
Sigma = jnp.array([
[1., 0., .8, 0.],
[0., 1., 0., .8],
[.8, 0., 1., 0.],
[0., .8, 0., 1.]
])
def logdensity(x, y):
return logdensity_fn(x, y, Sigma=Sigma)
MWG Sampling in BlackJAX#
In this case the conditional distributions \(p(\xx \mid \yy)\) and \(p(\yy \mid \xx)\) can be drawn from analytically (they are normal distributions). However, for illustrative purposes we’ll use an MCMC kernel to draw from each. Specifically, we’ll use an RMH kernel to draw from \(p(\xx \mid \yy)\) and an HMC kernel to draw from \(p(\yy \mid \xx)\). To implement the corresponding MWG algorithm in BlackJAX, we’ll write an mwg_kernel() for the problem which will do the following:
Maintain separate MCMC kernels to update each component of \(p(\xx, \yy)\) while holding the other fixed.
Apply the kernel updates correctly.
The issue with (2) is that each kernel update for a given MCMC Algorithm in BlackJAX refers to an algorithm-specific AlgorithmState. For example, RWState is a typing.NamedTuple class containing elements position and log_probability. In our MWG sampling problem at the beginning of step \(t\), RWState.log_probability will consist of \(\log p(\xx_{t-1}, \yy_{t-1})\). After updating \(\xx\), it will consist of \(\log p(\xx_{t}, \yy_{t-1})\). This happens automatically when we call blackjax.rmh.build_kernel(). However, after updating \(\yy\) (via HMC), we must manually update RWState.log_probability to consist of \(\log p(\xx_{t}, \yy_{t})\).
A general way of performing this manual update is to use the blackjax.algorithm.init() function of the given component’s MCMC algorithm to update the AlgorithmState. This function has arguments position and logdensity_fn. For example with the HMC component, after obtaining \(\xx_t\) but before drawing \(\yy_t\), the position would be \(\yy_{t-1}\) and the logdensity_fn function would be \(\log p(\xx_t, \cdot )\).
Using this approach, we now are now ready to implement the Gibbs sampling kernel in the code below.
Construct the MWG Kernel#
# MCMC initializers for each set of paramters
mwg_init_x = blackjax.rmh.init
mwg_init_y = blackjax.hmc.init
# MCMC updaters
mwg_step_fn_x = blackjax.rmh.build_kernel()
mwg_step_fn_y = blackjax.hmc.build_kernel() # default integrator, etc.
def mwg_kernel(rng_key, state, parameters):
"""
MWG kernel with RMH for ``x ~ p(x | y)`` and HMC for ``y ~ p(y | x)``.
Parameters
----------
rng_key
The PRNG key.
state
Dictionary with elements `x` and `y`, where the former is an ``RMCState`` object
and the latter is an ``HMCState`` object.
parameters
Dictionary with elements `x` and `y`, each of which is a dictionary of the parameters
to the corresponding algorithm's ``step_fn()``.
Returns
-------
Dictionary containing the updated ``state``.
"""
rng_key_x, rng_key_y = jax.random.split(rng_key, num=2)
# avoid modifying argument state as JAX functions should be pure
state = state.copy()
# --- update for x ---
# conditional logdensity of x given y
def logdensity_x(x): return logdensity(x=x, y=state["y"].position)
# give state["x"] the right log_density
state["x"] = mwg_init_x(
position=state["x"].position,
logdensity_fn=logdensity_x
)
# update state["x"]
state["x"], _ = mwg_step_fn_x(
rng_key=rng_key_x,
state=state["x"],
logdensity_fn=logdensity_x,
**parameters["x"]
)
# --- update for y ---
# conditional logdensity of y given x
def logdensity_y(y): return logdensity(y=y, x=state["x"].position)
# give state["y"] the right log_density
state["y"] = mwg_init_y(
position=state["y"].position,
logdensity_fn=logdensity_y
)
# update state["y"]
state["y"], _ = mwg_step_fn_y(
rng_key=rng_key_y,
state=state["y"],
logdensity_fn=logdensity_y,
**parameters["y"]
)
return state
Sampler Parameters#
parameters = {
"x": {
"transition_generator": blackjax.mcmc.random_walk.normal(.2 * jnp.eye(2))
},
"y": {
"inverse_mass_matrix": jnp.array([1., 1.]),
"num_integration_steps": 100,
"step_size": 1e-2
}
}
Set the Initial State of Each Algorithm#
initial_state = {
"x": mwg_init_x(
position=jnp.array([0., 0.]),
logdensity_fn=lambda x: logdensity(x=x, y=jnp.array([0., 0.]))
),
"y": mwg_init_y(
position=jnp.array([0., 0.]),
logdensity_fn=lambda y: logdensity(y=y, x=jnp.array([0., 0.]))
)
}
Build the Sampling Loop#
def sampling_loop(rng_key, initial_state, parameters, num_samples):
@jax.jit
def one_step(state, rng_key):
state = mwg_kernel(
rng_key=rng_key,
state=state,
parameters=parameters
)
positions = {k: state[k].position for k in state.keys()}
return state, positions
keys = jax.random.split(rng_key, num_samples)
_, positions = jax.lax.scan(one_step, initial_state, keys)
return positions
Sampling#
%%time
rng_key, sample_key = jax.random.split(rng_key)
positions = sampling_loop(sample_key, initial_state, parameters, 10_000)
CPU times: user 1.22 s, sys: 33.5 ms, total: 1.25 s
Wall time: 636 ms
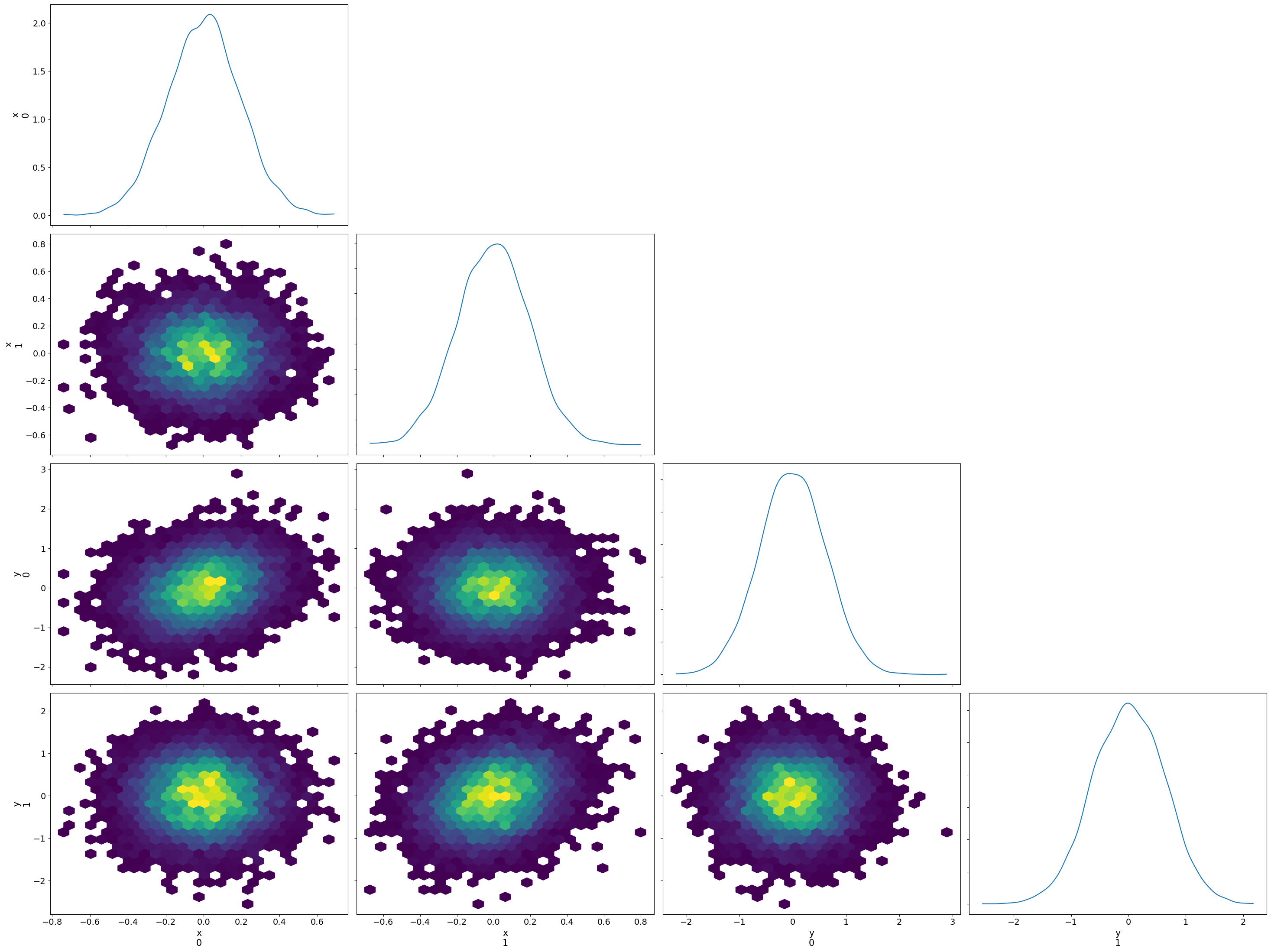
import matplotlib.pyplot as plt
import arviz as az
idata = az.from_dict(posterior={k: v[None, ...] for k, v in positions.items()})
az.plot_pair(idata, kind='hexbin', marginals=True)
plt.tight_layout();
/opt/hostedtoolcache/Python/3.12.12/x64/lib/python3.12/site-packages/arviz/__init__.py:39: FutureWarning:
ArviZ is undergoing a major refactor to improve flexibility and extensibility while maintaining a user-friendly interface.
Some upcoming changes may be backward incompatible.
For details and migration guidance, visit: https://python.arviz.org/en/latest/user_guide/migration_guide.html
warn(
General MWG Kernel#
The following code attempts to generalize the mwg_kernel() above to an arbitrary number of components \(p(\xx_1, \ldots, \xx_n)\).
def mwg_kernel_general(rng_key, state, logdensity_fn, step_fn, init, parameters):
"""
General MWG kernel.
Updates each component of ``state`` conditioned on all the others using a component-specific MCMC algorithm
Parameters
----------
rng_key
The PRNG key.
state
Dictionary where each item is the state of an MCMC algorithm, i.e., an object of type ``AlgorithmState``.
logdensity_fn
The log-density function on all components, where the arguments are the keys of ``state``.
step_fn
Dictionary with the same keys as ``state``,
each element of which is an MCMC stepping functions on the corresponding component.
init
Dictionary with the same keys as ``state``,
each elemtn of chi is an MCMC initializer corresponding to the stepping functions in `step_fn`.
parameters
Dictionary with the same keys as ``state``, each of which is a dictionary of parameters to
the MCMC algorithm for the corresponding component.
Returns
-------
Dictionary containing the updated ``state``.
"""
rng_keys = jax.random.split(rng_key, num=len(state))
rng_keys = dict(zip(state.keys(), rng_keys))
# avoid modifying argument state as JAX functions should be pure
state = state.copy()
for k in state.keys():
# logdensity of component k conditioned on all other components in state
def logdensity_k(value):
kwargs = {_k: state[_k].position for _k in state.keys()}
kwargs[k] = value
return logdensity_fn(**kwargs)
# give state[k] the right log_density
state[k] = init[k](
position=state[k].position,
logdensity_fn=logdensity_k
)
# update state[k]
state[k], _ = step_fn[k](
rng_key=rng_keys[k],
state=state[k],
logdensity_fn=logdensity_k,
**parameters[k]
)
return state
Build the Sampling Loop#
def sampling_loop_general(rng_key, initial_state, logdensity_fn, step_fn, init, parameters, num_samples):
@jax.jit
def one_step(state, rng_key):
state = mwg_kernel_general(
rng_key=rng_key,
state=state,
logdensity_fn=logdensity_fn,
step_fn=step_fn,
init=init,
parameters=parameters
)
positions = {k: state[k].position for k in state.keys()}
return state, positions
keys = jax.random.split(rng_key, num_samples)
_, positions = jax.lax.scan(one_step, initial_state, keys)
return positions
Sampling#
%%time
positions_general = sampling_loop_general(
rng_key=sample_key, # reuse PRNG key from above
initial_state=initial_state,
logdensity_fn=logdensity,
step_fn={
"x": mwg_step_fn_x,
"y": mwg_step_fn_y
},
init={
"x": mwg_init_x,
"y": mwg_init_y
},
parameters=parameters,
num_samples=10_000
)
CPU times: user 834 ms, sys: 14 ms, total: 848 ms
Wall time: 400 ms
Check Result#
jax.tree.map(lambda x, y: jnp.max(jnp.abs(x-y)), positions, positions_general)
{'x': Array(0., dtype=float32), 'y': Array(0., dtype=float32)}
Developer Notes#
The update method above (using
blackjax.algorithm.init()) should work out-of-the-box for most (if not all) MCMC algorithms in BlackJAX. However, it is not optimally efficient. For example for the RMH update, after obtaining \(\yy_{t-1}\) but before drawing \(\xx_t\), the method above would calculateRWState.log_densityto be \(\log p(\xx_{t-1}, \yy_{t-1})\). But we’ve already calculated this value from the previous HMC update of \(\yy_{t-1} \sim p(\yy \mid \xx_{t-1})\). So, we could save ourselves the cost of calculating the log-density twice, at the expense of a deeper understanding of the low-level components of the algorithms at hand and less generalizable code.The general MWG kernel prototyped above should be adequate for problems with a small number of components. However, the for-loop over the components of
stategets unrolled by the JAX JIT compiler (as discussed here), which can cause long compilation times when the number of components is large. To mitigate this problem, the for-loop could be replaced by alax.scan()primitive. For the sake of simplicity this approach is not fully developed here.
