Use Tempered SMC to Improve Exploration of MCMC Methods.#
Multimodal distributions are typically hard to sample from, in particular using energy based methods such as HMC, as you need high energy levels to escape a potential well.
Tempered SMC helps with this by considering a sequence of distributions:
where the tempering parameter \(\lambda_k\) takes increasing values between \(0\) and \(1\). Tempered SMC will also particularly shine when the MCMC step is not well calibrated (too small step size, etc) like in the example below.
Imports#
import jax
from datetime import date
rng_key = jax.random.key(int(date.today().strftime("%Y%m%d")))
import numpy as np
import jax.numpy as jnp
from jax.scipy.stats import multivariate_normal
import blackjax
import blackjax.smc.resampling as resampling
from blackjax.smc import extend_params
Sampling From a Bimodal Potential#
Experimental Setup#
We consider a prior distribution
and a potential function
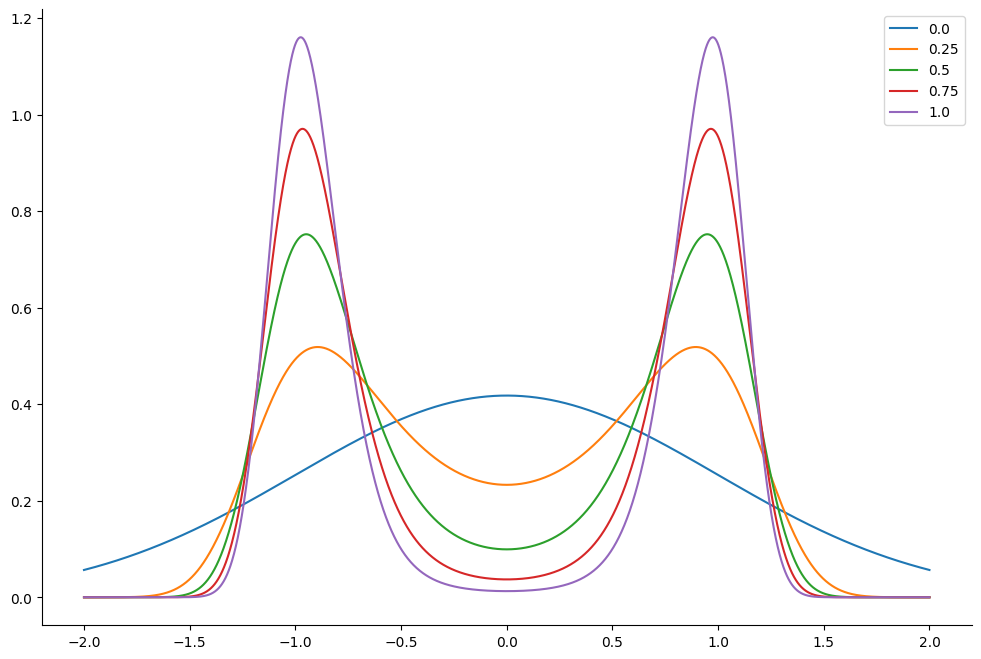
This corresponds to the following distribution. We plot the resulting tempered density for 5 different values of \(\lambda_k\) : from \(\lambda_k =1\) which correponds to the original density to \(\lambda_k=0\). The lower the value of \(\lambda_k\) the easier it is for the sampler to jump between the modes of the posterior density.
def V(x):
return 5 * jnp.square(jnp.sum(x**2, axis=-1) - 1)
def prior_log_prob(x):
d = x.shape[-1]
return multivariate_normal.logpdf(x, jnp.zeros((d,)), jnp.eye(d))
linspace = jnp.linspace(-2, 2, 5000)[..., None]
lambdas = jnp.linspace(0.0, 1.0, 5)
prior_logvals = prior_log_prob(linspace)
potential_vals = V(linspace)
log_res = prior_logvals - lambdas[..., None] * potential_vals
density = jnp.exp(log_res)
normalizing_factor = jnp.sum(density, axis=1, keepdims=True) * (
linspace[1] - linspace[0]
)
density /= normalizing_factor
<matplotlib.legend.Legend at 0x7ffb24271050>
def inference_loop(rng_key, mcmc_kernel, initial_state, num_samples):
@jax.jit
def one_step(state, k):
state, _ = mcmc_kernel(k, state)
return state, state
keys = jax.random.split(rng_key, num_samples)
_, states = jax.lax.scan(one_step, initial_state, keys)
return states
def full_logdensity(x):
return -V(x) + prior_log_prob(x)
inv_mass_matrix = jnp.eye(1)
n_samples = 10_000
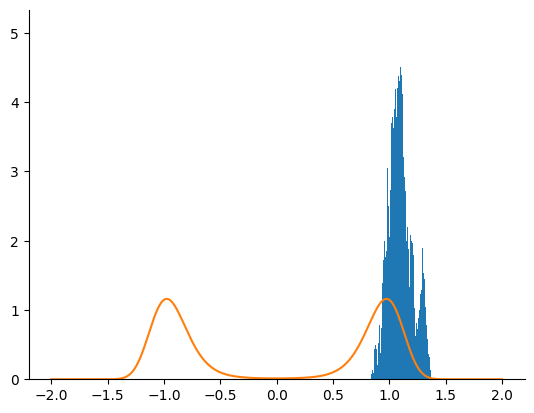
Sample with HMC#
We first try to sample from the posterior density using an HMC kernel.
%%time
hmc_parameters = dict(
step_size=1e-4, inverse_mass_matrix=inv_mass_matrix, num_integration_steps=50
)
hmc = blackjax.hmc(full_logdensity, **hmc_parameters)
hmc_state = hmc.init(jnp.ones((1,)))
rng_key, sample_key = jax.random.split(rng_key)
hmc_samples = inference_loop(sample_key, hmc.step, hmc_state, n_samples)
CPU times: user 2.14 s, sys: 108 ms, total: 2.24 s
Wall time: 1.45 s
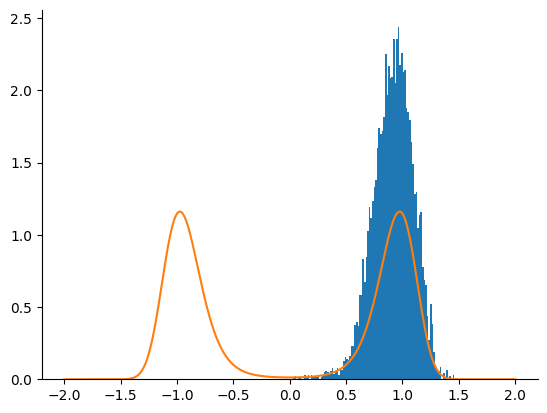
Sample with NUTS#
We now use a NUTS kernel.
%%time
nuts_parameters = dict(step_size=1e-4, inverse_mass_matrix=inv_mass_matrix)
nuts = blackjax.nuts(full_logdensity, **nuts_parameters)
nuts_state = nuts.init(jnp.ones((1,)))
rng_key, sample_key = jax.random.split(rng_key)
nuts_samples = inference_loop(sample_key, nuts.step, nuts_state, n_samples)
CPU times: user 18.1 s, sys: 170 ms, total: 18.2 s
Wall time: 17.1 s
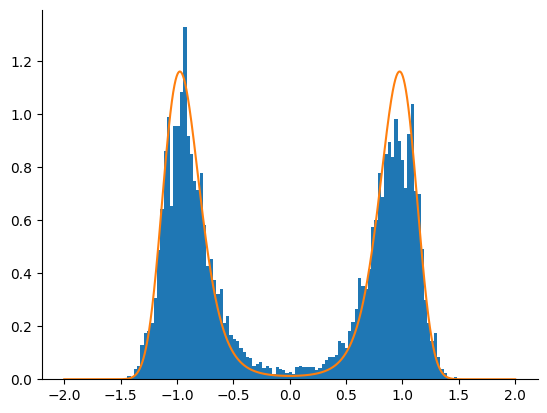
Tempered SMC with HMC Kernel#
We now use the adaptive tempered SMC algorithm with an HMC kernel. We only take one HMC step before resampling. The algorithm is run until \(\lambda_k\) crosses the \(\lambda_k = 1\) limit.
def smc_inference_loop(rng_key, smc_kernel, initial_state):
"""Run the temepered SMC algorithm.
We run the adaptive algorithm until the tempering parameter lambda reaches the value
lambda=1.
"""
def cond(carry):
i, state, _k = carry
return state.tempering_param < 1
def one_step(carry):
i, state, k = carry
k, subk = jax.random.split(k, 2)
state, _ = smc_kernel(subk, state)
return i + 1, state, k
n_iter, final_state, _ = jax.lax.while_loop(
cond, one_step, (0, initial_state, rng_key)
)
return n_iter, final_state
%%time
loglikelihood = lambda x: -V(x)
hmc_parameters = dict(
step_size=1e-4, inverse_mass_matrix=inv_mass_matrix, num_integration_steps=1
)
tempered = blackjax.adaptive_tempered_smc(
prior_log_prob,
loglikelihood,
blackjax.hmc.build_kernel(),
blackjax.hmc.init,
extend_params(hmc_parameters),
resampling.systematic,
0.5,
num_mcmc_steps=1,
)
rng_key, init_key, sample_key = jax.random.split(rng_key, 3)
initial_smc_state = jax.random.multivariate_normal(
init_key, jnp.zeros([1]), jnp.eye(1), (n_samples,)
)
initial_smc_state = tempered.init(initial_smc_state)
n_iter, smc_samples = smc_inference_loop(sample_key, tempered.step, initial_smc_state)
print("Number of steps in the adaptive algorithm: ", n_iter.item())
Number of steps in the adaptive algorithm: 12
CPU times: user 3.86 s, sys: 162 ms, total: 4.02 s
Wall time: 1.61 s
Sampling from the Rastrigin Potential#
Experimental Setup#
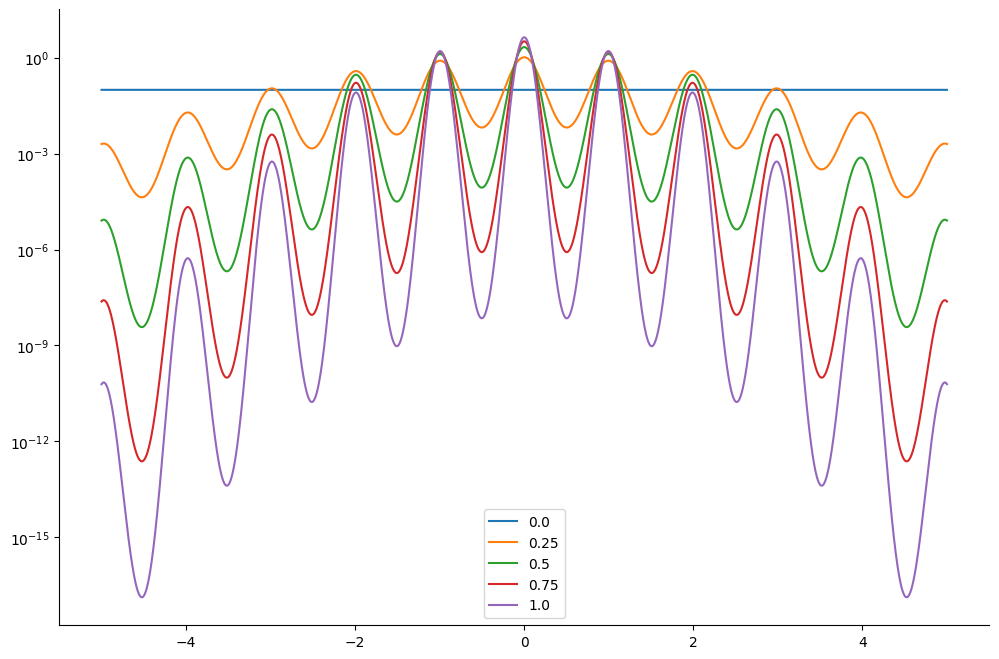
We consider a prior distribution \(p_0(x) = \mathcal{N}(x \mid 0_2, 2 I_2)\) and we want to sample from a Rastrigin type potential function \(V(x) = -2 A + \sum_{i=1}^2x_i^2 - A \cos(2 \pi x_i)\) where we choose \(A=10\). These potential functions are known to be particularly hard to sample.
We plot the resulting tempered density for 5 different values of \(\lambda_k\): from \(\lambda_k =1\) which correponds to the original density to \(\lambda_k=0\). The lower the value of \(\lambda_k\) the easier it is to sampler from the posterior log-density.
def V(x):
d = x.shape[-1]
res = -10 * d + jnp.sum(x**2 - 10 * jnp.cos(2 * jnp.pi * x), -1)
return res
linspace = jnp.linspace(-5, 5, 5000)[..., None]
lambdas = jnp.linspace(0.0, 1.0, 5)
potential_vals = V(linspace)
log_res = lambdas[..., None] * potential_vals
density = jnp.exp(-log_res)
normalizing_factor = jnp.sum(density, axis=1, keepdims=True) * (
linspace[1] - linspace[0]
)
density /= normalizing_factor
<matplotlib.legend.Legend at 0x7ffaeccecf10>
def inference_loop(rng_key, mcmc_kernel, initial_state, num_samples):
def one_step(state, k):
state, _ = mcmc_kernel(k, state)
return state, state
keys = jax.random.split(rng_key, num_samples)
_, states = jax.lax.scan(one_step, initial_state, keys)
return states
inv_mass_matrix = jnp.eye(1)
n_samples = 1_000
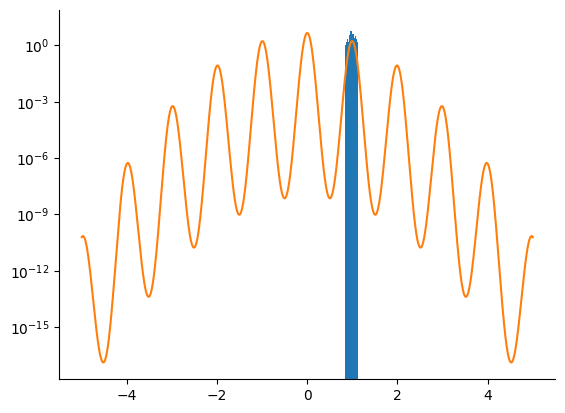
HMC Sampler#
We first try to sample from the posterior density using an HMC kernel.
%%time
loglikelihood = lambda x: -V(x)
hmc_parameters = dict(
step_size=1e-2, inverse_mass_matrix=inv_mass_matrix, num_integration_steps=50
)
hmc = blackjax.hmc(full_logdensity, **hmc_parameters)
hmc_state = hmc.init(jnp.ones((1,)))
rng_key, sample_key = jax.random.split(rng_key)
hmc_samples = inference_loop(sample_key, hmc.step, hmc_state, n_samples)
CPU times: user 990 ms, sys: 28.1 ms, total: 1.02 s
Wall time: 554 ms
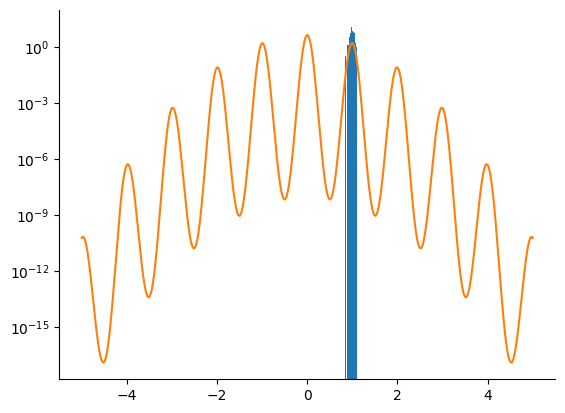
NUTS Sampler#
We do the same using a NUTS kernel.
%%time
nuts_parameters = dict(step_size=1e-2, inverse_mass_matrix=inv_mass_matrix)
nuts = blackjax.nuts(full_logdensity, **nuts_parameters)
nuts_state = nuts.init(jnp.ones((1,)))
rng_key, sample_key = jax.random.split(rng_key)
nuts_samples = inference_loop(sample_key, nuts.step, nuts_state, n_samples)
CPU times: user 1.44 s, sys: 45.8 ms, total: 1.48 s
Wall time: 770 ms
Tempered SMC with HMC Kernel#
We now use the adaptive tempered SMC algorithm with an HMC kernel. We only take one HMC step before resampling. The algorithm is run until \(\lambda_k\) crosses the \(\lambda_k = 1\) limit. We correct the bias introduced by the (arbitrary) prior.
%%time
loglikelihood = lambda x: -V(x)
hmc_parameters = dict(
step_size=1e-2, inverse_mass_matrix=inv_mass_matrix, num_integration_steps=100
)
tempered = blackjax.adaptive_tempered_smc(
prior_log_prob,
loglikelihood,
blackjax.hmc.build_kernel(),
blackjax.hmc.init,
extend_params(hmc_parameters),
resampling.systematic,
0.75,
num_mcmc_steps=1,
)
rng_key, init_key, sample_key = jax.random.split(rng_key, 3)
initial_smc_state = jax.random.multivariate_normal(
init_key, jnp.zeros([1]), jnp.eye(1), (n_samples,)
)
initial_smc_state = tempered.init(initial_smc_state)
n_iter, smc_samples = smc_inference_loop(sample_key, tempered.step, initial_smc_state)
print("Number of steps in the adaptive algorithm: ", n_iter.item())
Number of steps in the adaptive algorithm: 8
CPU times: user 3.21 s, sys: 81.9 ms, total: 3.3 s
Wall time: 1.37 s
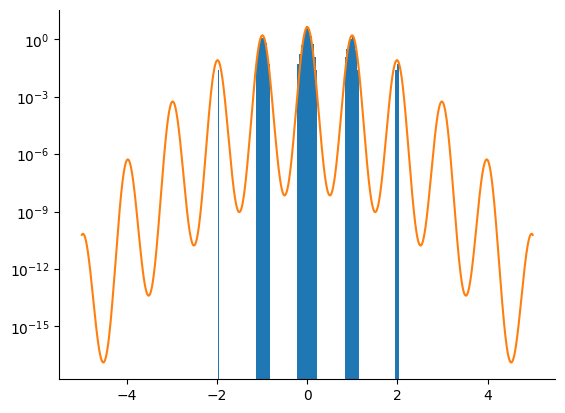
The tempered SMC algorithm with the HMC kernel clearly outperfoms the HMC and NUTS kernels alone.